Downloading MODIS data
MODIS data can be acquired through multiple services, and it can be confusing to find out what to get where. This website has a lot of information on MODIS resources.
Websites are great for preliminary data exploration, they are not ideal
for accessing large number of files or when automatic updates are
required. Manually downloading also makes workflows less reproducible.
So once you know what data you need, it is better to use automatic
downloading methods. Here we show how to use R package luna to
download MODIS data for a geographic area and time period.
A large number of MODIS products is available. We first need to find data for the product that we deem best suited for our study.
library(terra)
library(luna)
# lists all products that are currently searchable0
prod <- getProducts()
head(prod)
## provider concept_id short_name version
## 1 GHRC C1000-GHRC dc8capac 1
## 5 CDDIS C1000000000-CDDIS CDDIS_DORIS_data_cycle 1
## 8 SEDAC C1000000000-SEDAC CIESIN_SEDAC_EPI_2012 2012.00
## 12 CDDIS C1000000001-CDDIS CDDIS_DORIS_data_rinex 1
## 15 LARC C1000000001-LARC MI3QRDF 1
## 22 SEDAC C1000000001-SEDAC CIESIN_SEDAC_FERMANv1_NMAN 1.00
# to find the MODIS products
modis <- getProducts("^MOD|^MYD|^MCD")
head(modis)
## provider concept_id short_name version
## 409 LPDAAC_ECS C1000000120-LPDAAC_ECS MOD44B 051
## 837 LPDAAC_ECS C1000000400-LPDAAC_ECS MCD43D10 006
## 848 LPDAAC_ECS C1000000401-LPDAAC_ECS MCD43D33 006
## 851 LPDAAC_ECS C1000000402-LPDAAC_ECS MCD43D45 006
## 854 LPDAAC_ECS C1000000403-LPDAAC_ECS MCD43D26 006
## 857 LPDAAC_ECS C1000000404-LPDAAC_ECS MCD43D49 006
We will use “MOD09A1” for this turorial.
product <- "MOD09A1"
To learn more about a specific product you can launch a webpage
productInfo(product)
Note that the entire MODIS archive is regularly re-processed for overall imporvement and revisions. We use version 6 or later for our analysis.
Once we finalize the product we want to use, we define some parameters for the data we want: product name, start and end date, and area of interest.
start <- "2010-01-01"
end <- "2010-01-07"
We will download an example MODIS 8-day composite tile. Our area of interest is Marsabit county, Kenya. To define the area of interest, we can define a spatial extent, or use an object that has an extent. Here we use a polygon for Marsabit.
ken <- geodata::gadm("Kenya", level=1, path=".")
ken
## class : SpatVector
## geometry : polygons
## dimensions : 47, 11 (geometries, attributes)
## extent : 33.90959, 41.92622, -4.720417, 5.061166 (xmin, xmax, ymin, ymax)
## coord. ref. : lon/lat WGS 84 (EPSG:4326)
## names : GID_1 GID_0 COUNTRY NAME_1 VARNAME_1 NL_NAME_1 TYPE_1
## type : <chr> <chr> <chr> <chr> <chr> <chr> <chr>
## values : KEN.1_1 KEN Kenya Baringo NA NA County
## KEN.2_1 KEN Kenya Bomet NA NA County
## KEN.3_1 KEN Kenya Bungoma NA NA County
## ENGTYPE_1 CC_1 HASC_1 ISO_1
## <chr> <chr> <chr> <chr>
## County 30 KE.BA KE-01
## County 36 KE.BO KE-02
## County 39 KE.BN KE-03
ken is a SpatVector of polygons. We can subset it get the
polygon for Marsabit:
i <- ken$NAME_1 == "Marsabit"
aoi <- ken[i,]
And the check our results we make a map
plot(ken, col="light gray")
lines(aoi, col="red", lwd=2)
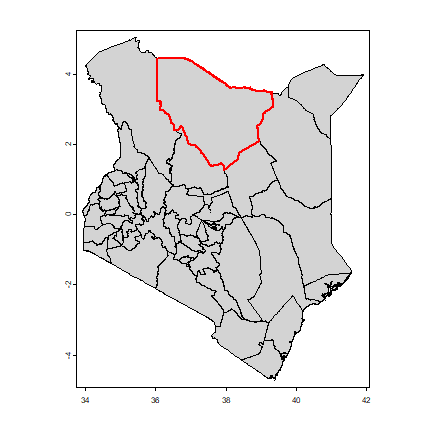
Let’s now find out what MODIS data is available for this area. We can search the data available from a NASA server
mf <- luna::getNASA(product, start, end, aoi=aoi, download = FALSE)
mf
## [1] "MOD09A1.A2009361.h21v08.061.2021149144347.hdf"
## [2] "MOD09A1.A2010001.h21v08.061.2021150093948.hdf"
To download the tiles, usually you would download them to a folder where you save the data for your project. Here we use the temporary directory. You should use a specific direcory of your choice instead.
datadir <- file.path(dirname(tempdir()), "_modis")
dir.create(datadir, showWarnings=FALSE)
You also need to provide the username and password for your (free) EOSDIS account. If you do not have an account, you can sign up here. My passwords are stored in a file that I read below (sorry, I cannot show you the values).
up <- readRDS("../../../../pwds.rds")
up <- up[up$service == "EOSDIS", ]
Now we are ready to download the data
mf <- luna::getNASA(product, start, end, aoi=aoi, download=TRUE, overwrite=TRUE,
path=datadir, username=up$user, password=up$pwd)
## Downloading: 27 B Downloading: 27 B Downloading: 27 B Downloading: 320 B Downloading: 320 B Downloading: 320 B | | | 0% | | | 1% | |= | 1% | |= | 2% | |== | 2% | |== | 3% | |== | 4% | |=== | 4% | |=== | 5% | |==== | 5% | |==== | 6% | |===== | 6% | |===== | 7% | |===== | 8% | |====== | 8% | |====== | 9% | |======= | 9% | |======= | 10% | |======= | 11% | |======== | 11% | |======== | 12% | |========= | 12% | |========= | 13% | |========= | 14% | |========== | 14% | |========== | 15% | |=========== | 15% | |=========== | 16% | |============ | 16% | |============ | 17% | |============ | 18% | |============= | 18% | |============= | 19% | |============== | 19% | |============== | 20% | |============== | 21% | |=============== | 21% | |=============== | 22% | |================ | 22% | |================ | 23% | |================ | 24% | |================= | 24% | |================= | 25% | |================== | 25% | |================== | 26% | |=================== | 26% | |=================== | 27% | |=================== | 28% | |==================== | 28% | |==================== | 29% | |===================== | 29% | |===================== | 30% | |===================== | 31% | |====================== | 31% | |====================== | 32% | |======================= | 32% | |======================= | 33% | |======================= | 34% | |======================== | 34% | |======================== | 35% | |========================= | 35% | |========================= | 36% | |========================== | 36% | |========================== | 37% | |========================== | 38% | |=========================== | 38% | |=========================== | 39% | |============================ | 39% | |============================ | 40% | |============================ | 41% | |============================= | 41% | |============================= | 42% | |============================== | 42% | |============================== | 43% | |============================== | 44% | |=============================== | 44% | |=============================== | 45% | |================================ | 45% | |================================ | 46% | |================================= | 46% | |================================= | 47% | |================================= | 48% | |================================== | 48% | |================================== | 49% | |=================================== | 49% | |=================================== | 50% | |=================================== | 51% | |==================================== | 51% | |==================================== | 52% | |===================================== | 52% | |===================================== | 53% | |===================================== | 54% | |====================================== | 54% | |====================================== | 55% | |======================================= | 55% | |======================================= | 56% | |======================================== | 56% | |======================================== | 57% | |======================================== | 58% | |========================================= | 58% | |========================================= | 59% | |========================================== | 59% | |========================================== | 60% | |========================================== | 61% | |=========================================== | 61% | |=========================================== | 62% | |============================================ | 62% | |============================================ | 63% | |============================================ | 64% | |============================================= | 64% | |============================================= | 65% | |============================================== | 65% | |============================================== | 66% | |=============================================== | 66% | |=============================================== | 67% | |=============================================== | 68% | |================================================ | 68% | |================================================ | 69% | |================================================= | 69% | |================================================= | 70% | |================================================= | 71% | |================================================== | 71% | |================================================== | 72% | |=================================================== | 72% | |=================================================== | 73% | |=================================================== | 74% | |==================================================== | 74% | |==================================================== | 75% | |===================================================== | 75% | |===================================================== | 76% | |====================================================== | 76% | |====================================================== | 77% | |====================================================== | 78% | |======================================================= | 78% | |======================================================= | 79% | |======================================================== | 79% | |======================================================== | 80% | |======================================================== | 81% | |========================================================= | 81% | |========================================================= | 82% | |========================================================== | 82% | |========================================================== | 83% | |========================================================== | 84% | |=========================================================== | 84% | |=========================================================== | 85% | |============================================================ | 85% | |============================================================ | 86% | |============================================================= | 86% | |============================================================= | 87% | |============================================================= | 88% | |============================================================== | 88% | |============================================================== | 89% | |=============================================================== | 89% | |=============================================================== | 90% | |=============================================================== | 91% | |================================================================ | 91% | |================================================================ | 92% | |================================================================= | 92% | |================================================================= | 93% | |================================================================= | 94% | |================================================================== | 94% | |================================================================== | 95% | |=================================================================== | 95% | |=================================================================== | 96% | |==================================================================== | 96% | |==================================================================== | 97% | |==================================================================== | 98% | |===================================================================== | 98% | |===================================================================== | 99% | |======================================================================| 99% | |======================================================================| 100%
## | | | 0% | | | 1% | |= | 1% | |= | 2% | |== | 2% | |== | 3% | |== | 4% | |=== | 4% | |=== | 5% | |==== | 5% | |==== | 6% | |===== | 6% | |===== | 7% | |===== | 8% | |====== | 8% | |====== | 9% | |======= | 9% | |======= | 10% | |======= | 11% | |======== | 11% | |======== | 12% | |========= | 12% | |========= | 13% | |========= | 14% | |========== | 14% | |========== | 15% | |=========== | 15% | |=========== | 16% | |============ | 16% | |============ | 17% | |============ | 18% | |============= | 18% | |============= | 19% | |============== | 19% | |============== | 20% | |============== | 21% | |=============== | 21% | |=============== | 22% | |================ | 22% | |================ | 23% | |================ | 24% | |================= | 24% | |================= | 25% | |================== | 25% | |================== | 26% | |=================== | 26% | |=================== | 27% | |=================== | 28% | |==================== | 28% | |==================== | 29% | |===================== | 29% | |===================== | 30% | |===================== | 31% | |====================== | 31% | |====================== | 32% | |======================= | 32% | |======================= | 33% | |======================= | 34% | |======================== | 34% | |======================== | 35% | |========================= | 35% | |========================= | 36% | |========================== | 36% | |========================== | 37% | |========================== | 38% | |=========================== | 38% | |=========================== | 39% | |============================ | 39% | |============================ | 40% | |============================ | 41% | |============================= | 41% | |============================= | 42% | |============================== | 42% | |============================== | 43% | |============================== | 44% | |=============================== | 44% | |=============================== | 45% | |================================ | 45% | |================================ | 46% | |================================= | 46% | |================================= | 47% | |================================= | 48% | |================================== | 48% | |================================== | 49% | |=================================== | 49% | |=================================== | 50% | |=================================== | 51% | |==================================== | 51% | |==================================== | 52% | |===================================== | 52% | |===================================== | 53% | |===================================== | 54% | |====================================== | 54% | |====================================== | 55% | |======================================= | 55% | |======================================= | 56% | |======================================== | 56% | |======================================== | 57% | |======================================== | 58% | |========================================= | 58% | |========================================= | 59% | |========================================== | 59% | |========================================== | 60% | |========================================== | 61% | |=========================================== | 61% | |=========================================== | 62% | |============================================ | 62% | |============================================ | 63% | |============================================ | 64% | |============================================= | 64% | |============================================= | 65% | |============================================== | 65% | |============================================== | 66% | |=============================================== | 66% | |=============================================== | 67% | |=============================================== | 68% | |================================================ | 68% | |================================================ | 69% | |================================================= | 69% | |================================================= | 70% | |================================================= | 71% | |================================================== | 71% | |================================================== | 72% | |=================================================== | 72% | |=================================================== | 73% | |=================================================== | 74% | |==================================================== | 74% | |==================================================== | 75% | |===================================================== | 75% | |===================================================== | 76% | |====================================================== | 76% | |====================================================== | 77% | |====================================================== | 78% | |======================================================= | 78% | |======================================================= | 79% | |======================================================== | 79% | |======================================================== | 80% | |======================================================== | 81% | |========================================================= | 81% | |========================================================= | 82% | |========================================================== | 82% | |========================================================== | 83% | |========================================================== | 84% | |=========================================================== | 84% | |=========================================================== | 85% | |============================================================ | 85% | |============================================================ | 86% | |============================================================= | 86% | |============================================================= | 87% | |============================================================= | 88% | |============================================================== | 88% | |============================================================== | 89% | |=============================================================== | 89% | |=============================================================== | 90% | |=============================================================== | 91% | |================================================================ | 91% | |================================================================ | 92% | |================================================================= | 92% | |================================================================= | 93% | |================================================================= | 94% | |================================================================== | 94% | |================================================================== | 95% | |=================================================================== | 95% | |=================================================================== | 96% | |==================================================================== | 96% | |==================================================================== | 97% | |==================================================================== | 98% | |===================================================================== | 98% | |===================================================================== | 99% | |======================================================================| 99% | |======================================================================| 100%
basename(mf)
## [1] "MOD09A1.A2009361.h21v08.061.2021149144347.hdf"
## [2] "MOD09A1.A2010001.h21v08.061.2021150093948.hdf"
In the next chapters we will use the downloaded files.